Introduction
 The MassARRAY® System by Agena Bioscience is a multifaceted molecular research platform widely used in pharmacogenomics (PGx research), genotyping of SNPs and indels, CNV detection, and oncology research workflows. Powered by proprietary chemistries—iPLEX® Pro, iPLEX® HS, UltraSEEK®, and Agena ClearSEEK™—the MassARRAY® System provides flexible and targeted solutions for diverse genomics research applications.
The MassARRAY® System by Agena Bioscience is a multifaceted molecular research platform widely used in pharmacogenomics (PGx research), genotyping of SNPs and indels, CNV detection, and oncology research workflows. Powered by proprietary chemistries—iPLEX® Pro, iPLEX® HS, UltraSEEK®, and Agena ClearSEEK™—the MassARRAY® System provides flexible and targeted solutions for diverse genomics research applications.
Topics
- MassARRAY System Workflow & Chemistries Overview
- Genotyping and Sample Integrity with iPlex Pro Chemistry
- iPLEX Pro Chemistry & Workflow
- iPLEX Panels for Pharmacogenetics and Hereditary Genetics Genotyping
- iPLEX Panels for Sample Integrity and Validity
- Mutation profiling with iPLEX HS chemistry – for variant detection down to 1% VAF
- iPLEX HS Chemistry & Workflow
- Panels for Mutation Profiling
- Liquid biopsy with UltraSEEK chemistry – for variant detection down to 0.1% VAF
- UltraSEEK Chemistry & Workflow
- Panels for Liquid Biopsy
- Custom Panels
MassARRAY System Workflow & Flexible Chemistries
All chemistries on the MassARRAY System employ mass spectrometry and end-point PCR (polymerase chain reaction) analysis in research workflows. After nucleic acid extraction, samples are processed through a simple workflow:
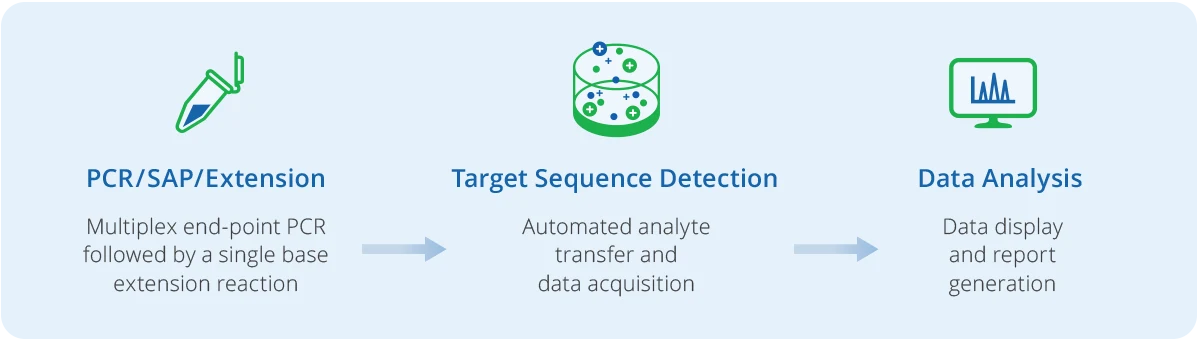
The MassARRAY System is powered by matrix-assisted laser desorption/ionization – time of flight (MALDI-TOF) for the identification of genetic variants. Each panel is designed to identify genetic variants according to their individual mass. DNA or cDNA is amplified using one of several Agena Bioscience chemistries, available through pre-designed or custom panels for research needs.
Genotyping and Sample Integrity with iPlex Pro Chemistry
iPLEX Pro Chemistry & Workflow
The robust multiplexed primer extension chemistry of the iPLEX assay combines with MALDI-TOF to analyze up to thousands of genotypes per day with a high level of stringency and reproducibility. iPLEX single-base extension chemistry enables highly reliable allele calling thresholds at high multiplexing levels.1
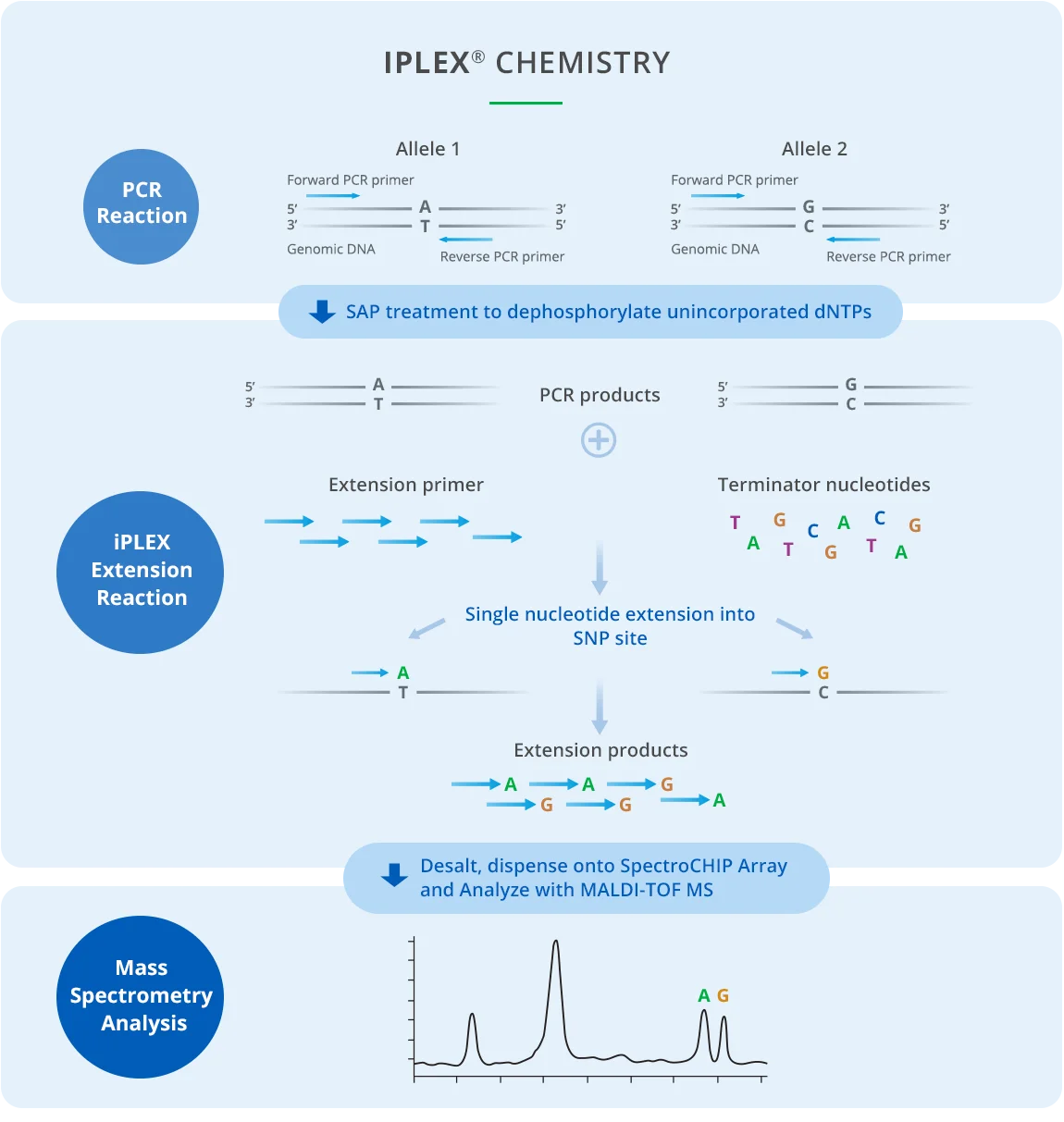
In the iPLEX process, genomic DNA or cDNA is amplified in 96- or 384-well microtiter plates using iPLEX reagents. Once PCR is complete, excess nucleotides are dephosphorylated by shrimp alkaline phosphatase (SAP). Next, the iPLEX single-base extension reaction uses a mix of oligonucleotide extension primers to anneal the amplified DNA, which is added with an extension enzyme and terminators. Extension primers anneal directly adjacent to each SNP (single nucleotide polymorphisms) site to be assayed, extended and terminated by a single complementary base at the genotyping target site. Analytes are desalted and transferred from the microtiter plate via an automated nanodispenser or manual dispensation onto a SpectroCHIP® Array, where they crystallize with a pre-spotted MALDI matrix.
After the process is complete, the SpectroCHIP Array is loaded into the MassARRAY Analyzer, which detects DNA within a mass range of approximately 4,500 Da to 9,000 Da and distinguishes between analytes separated by 16 Da. MassARRAY Typer software generates a report identifying homozygous and heterozygous SNP alleles.
iPLEX Pro Panels for Pharmacogenetics and Hereditary Genetics Genotyping
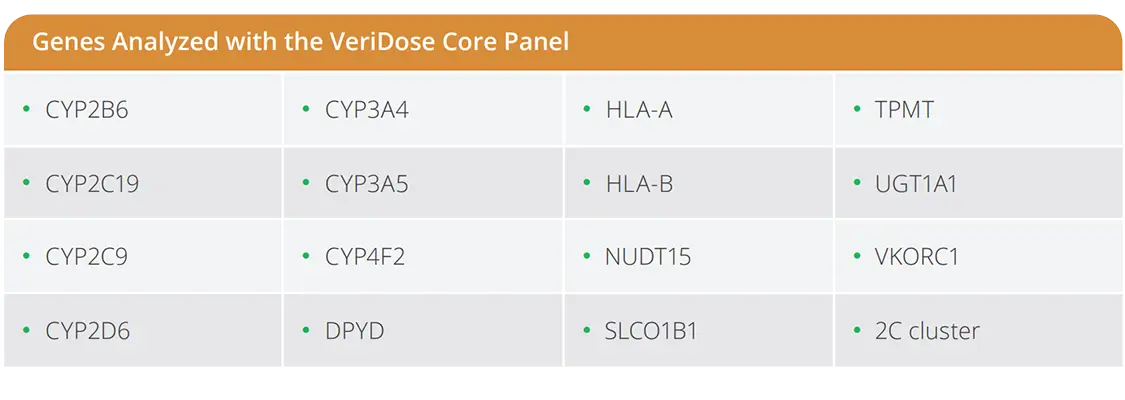
The VeriDose® Core Panel targets the most relevant pharmacogenetic variants across 16 key drug metabolism genes, delivering genotype data for 85 society-recommended SNPs/INDELs. Key features include:
- Focused content: 85 SNPs/INDELs across 16 genes recommended by leading PGx societies
- Efficient workflow: Run on the same plate as the VeriDose CYP2D6 CNV Panel
- Clear reports: Software delivers diplotype, haplotype, and CNV results
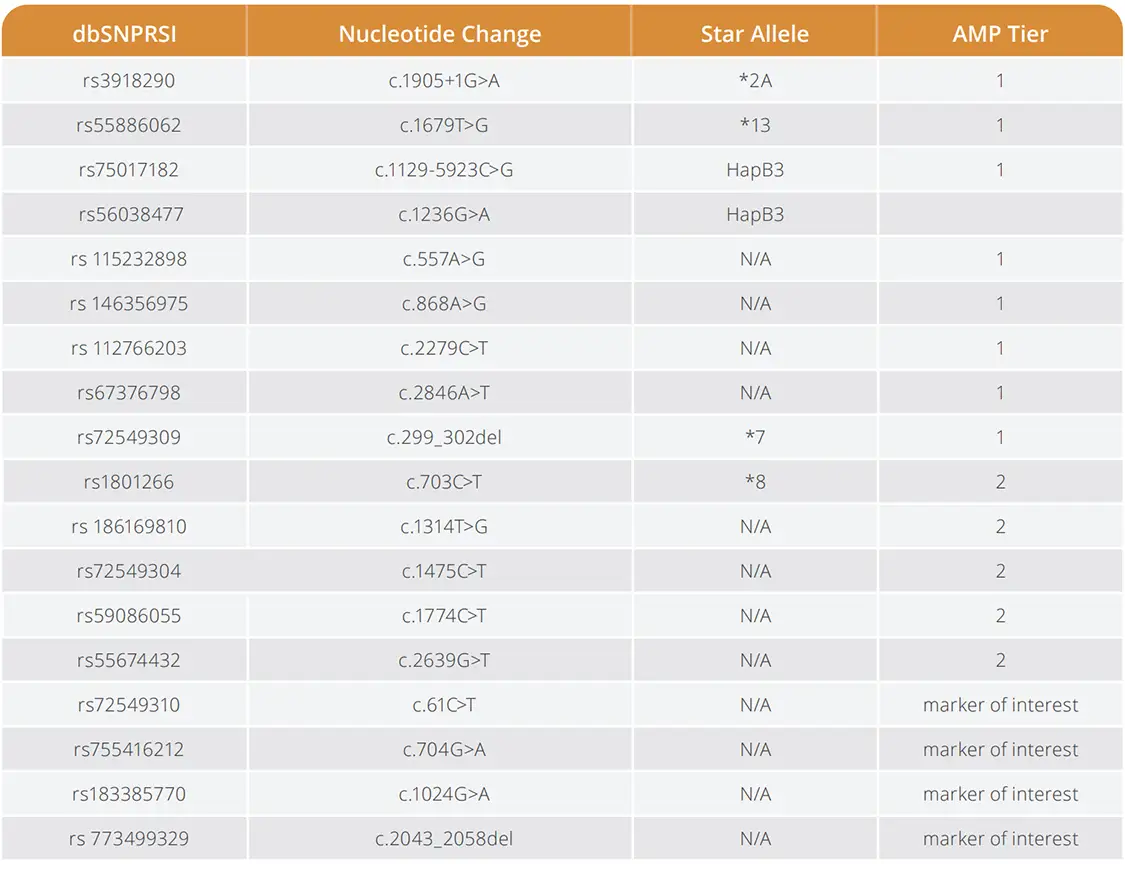
The VeriDose® DPYD Plus Panel delivers comprehensive DPYD variant coverage for pharmacogenomics research, including all AMP Tier 1 and Tier 2 variants plus four additional targets. Key features include:
- Comprehensive coverage: AMP Tier 1 & 2 variants plus four additional targets
- Cost efficiency: Targeted workflow with no minimum batch sizes and lower cost than NGS
- Simplified workflow: Single-well, plate-based setup with built-in analysis
- Scalable throughput: Run 1 to 1,000+ samples per day in 96- or 384-well formats
DPYD Assay Components:
The HFE Genotyping Panel supports hereditary hemochromatosis research across diverse laboratory settings. Key features include:
- 6-variant coverage: Recommended variants plus additional targets for broader population studies
- Streamlined workflow: Single-well multiplexing reduces hands-on time and errors
- Integrated analysis: Built-in software removes bioinformatics complexity
- Cost-effective: Lower cost per data point for labs running 100+ tests per month
HFE Assay Components
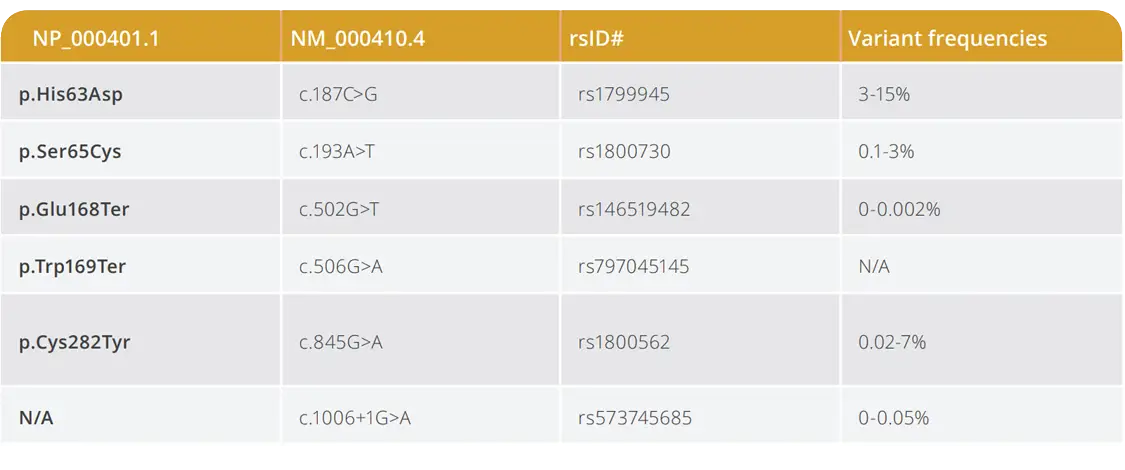
The CFTR 100+ Panel supports cystic fibrosis research by detecting 122 CFTR variants, including all 100 variants recommended by the American College of Medical Genetics and Genomics plus additional variants informed by emerging research and customer input.
iPLEX Pro Panels for Sample Integrity and Validity
Each of the iPLEX Pro Sample Integrity panels enables rapid and reproducible sample identification and DNA quality measurements, to generate a sample’s unique genetic fingerprint, which is used to deliver a “Match” or “Mismatch” result.
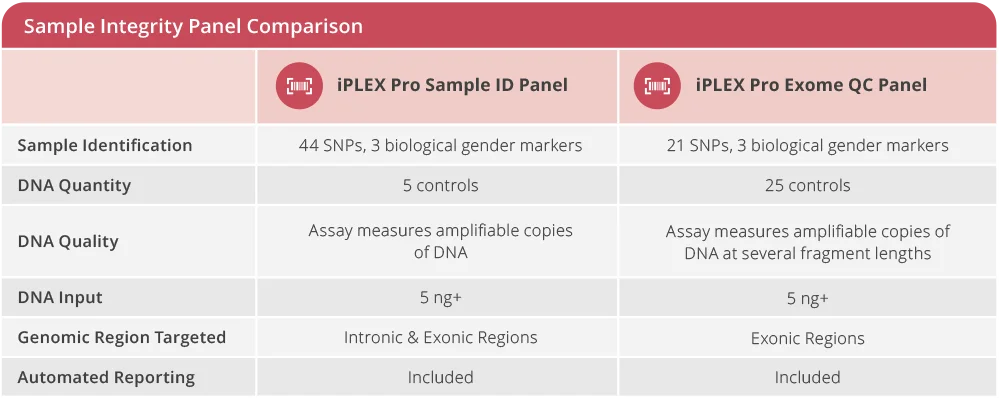
The iPLEX Pro Sample ID Panel is ideal for rapid sample management in research laboratories, biobanks, and toxicology workflows. This panel creates a unique genetic fingerprint for each subject, which is used to compare future samples to ensure the validity of the specimen or alert the laboratory that manipulation or mix-up has occurred.
The iPLEX Pro Exome QC Panel also determines a sample’s identity, DNA quality, and DNA quantity. It targets 21 SNPs (single nucleotide polymorphisms) in exonic regions of the genome and three biological gender markers. Additionally, it provides the number of intact, amplifiable DNA copies across a size range of 100 – 500 nucleotides.
Mutation profiling with iPLEX HS chemistry – for variant detection down to 1% VAF
iPLEX HS Chemistry & Workflow
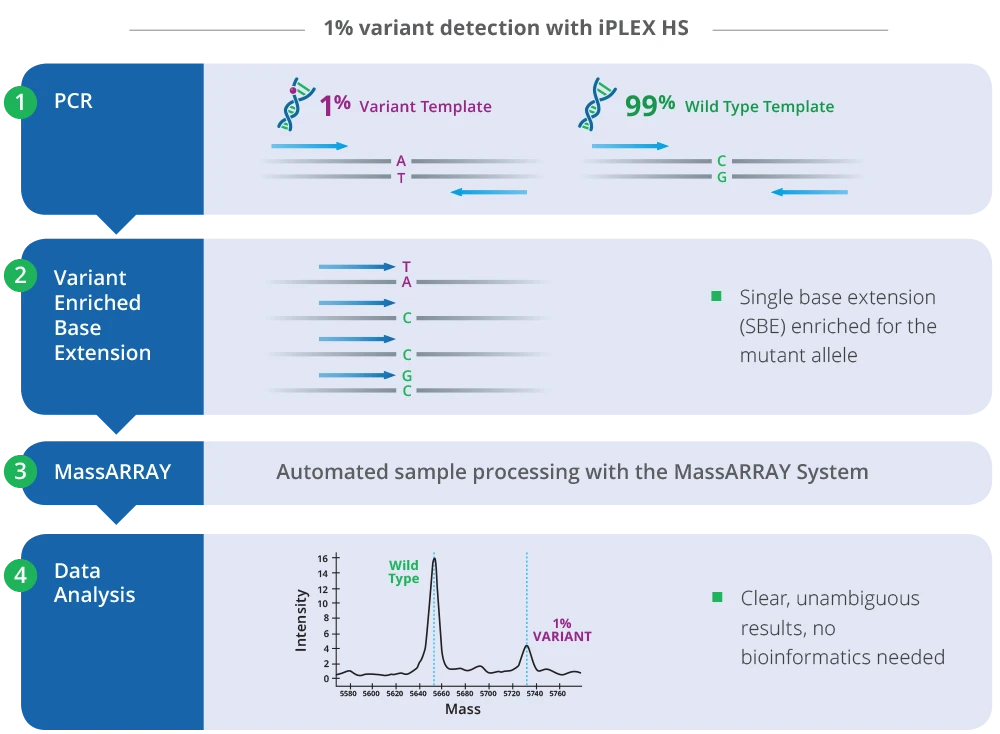
iPLEX HS panels on the MassARRAY System enable multiplexed detection of genetic variants present in research samples such as FFPE material. iPLEX HS uses single-base extension enriched for mutant alleles to identify variants of interest, while minimizing sample rejection, in as little as one hour of hands-on time and a total of eight hours for data generation.
iPLEX HS uses as little as 10 nanograms of input DNA with low-frequency variant detection for alleles as low as 1%. After PCR amplification, data output from the MassARRAY System distinguishes variants from wild-type signal. Clear spectra provide user-friendly, at-a-glance analysis of variants of interest. The short amplicon length (80–120 bp) makes it compatible with degraded samples.
Panels for Mutation Profiling
The iPLEX HS Lung Panel enables multiplexed variant detection in lung cancer research, featuring:
- Identification of 70 variants of research interest across BRAF, EGFR, ERBB2, KRAS, and PIK3CA
- Detection of variants at allele frequencies as low as 1%
- Use of as little as 10 ng of DNA
- Approximately 1 hour of hands-on time and 8 hours total time

The iPLEX HS Colon panel enables multiplexed variant detection in colorectal cancer research samples, featuring:
- Identification of more than 80 variants of research interest across BRAF, EGFR, KRAS, NRAS, PIK3CA, and other genes
- Detection of variants at allele frequencies as low as 1%
- Use of as little as 10 ng of DNA
- Approximately 1 hour of hands-on time and 8 hours total time

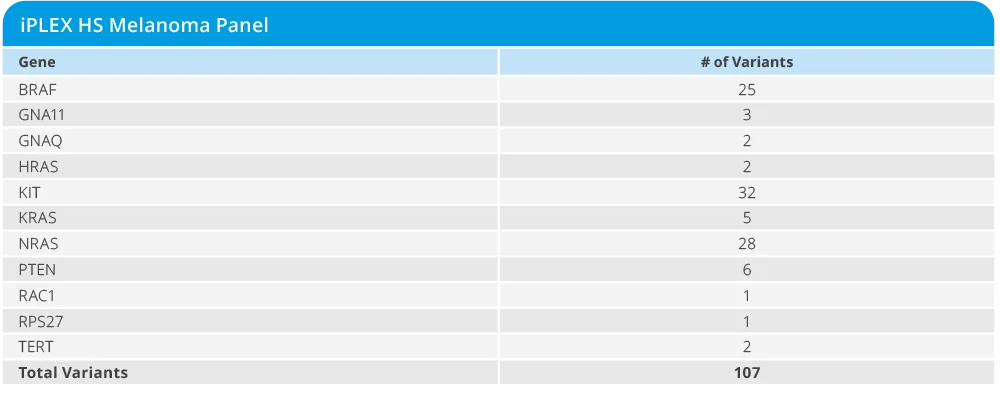
The iPLEX HS Melanoma panel enables multiplexed variant detection in melanoma research samples, featuring:
- Identification of more than 90 variants of research interest across BRAF, KIT, NRAS, TERT, and other genes
- Detection of variants at allele frequencies as low as 1%
- Use of as little as 10 ng of DNA
- Approximately 1 hour of hands-on time and 8 hours total time
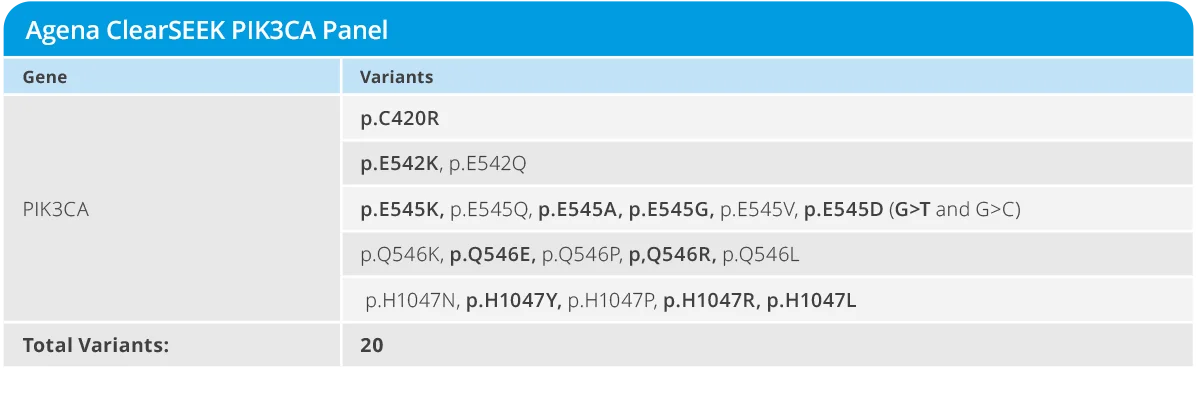
The PIK3CA Breast Panel targets a set of 20 variants in PIK3CA associated with response to targeted therapy, including the activating mutations recommended by the National Comprehensive Cancer Network Guidelines. Panel features include:
- Identification of 20 PIK3CA variants of research interest
- Identification of variants at as low as 1% variant allele frequency
- Use as little as 10 ng of DNA
- 1 hour of hands-on time and 8 hours of total time
Liquid biopsy with UltraSEEK chemistry – for variant detection down to 0.1% VAF
UltraSEEK Chemistry & Workflow
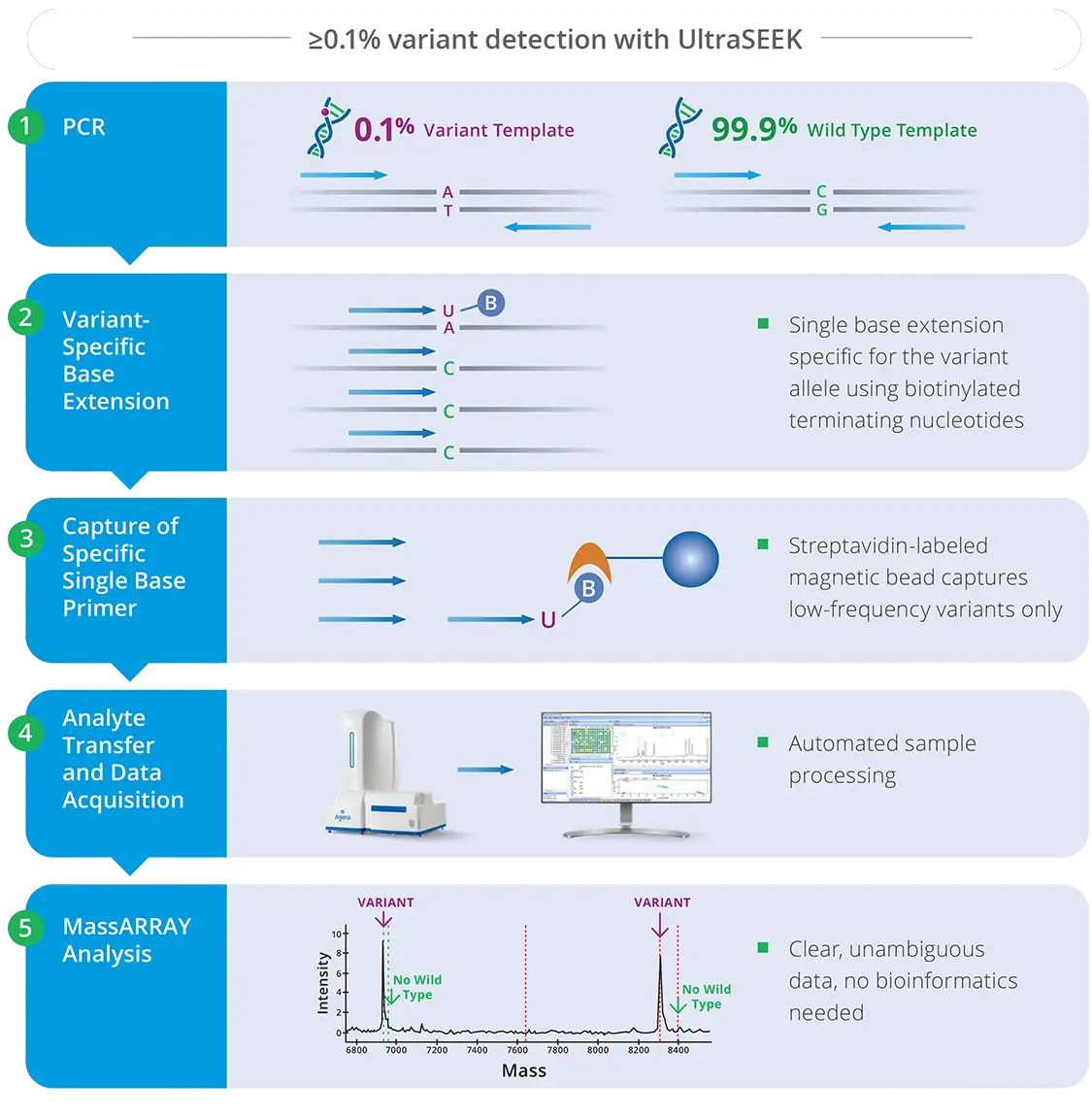
UltraSEEK chemistry on the MassARRAY System provides a targeted, multiplexed method for detecting rare events, enabling the identification of low-level genetic variants of research interest in oncology research samples, supporting studies of variant dynamics in model systems, and providing orthogonal validation of next-generation sequencing (NGS) or PCR-based data in a research context.
UltraSEEK chemistry consists of PCR amplification of a set of pre-defined loci harboring mutations of interest. This is followed by mutant-specific single-base extension and capture by streptavidin-labeled magnetic beads. Built-in controls verify the presence of DNA templates in the reaction.
Panels for Liquid Biopsy
UltraSEEK panels are available for multiple oncology research areas including:
The UltraSEEK Lung panel enables multiplexed variant detection in lung cancer research samples, featuring:
- Identification of more than 75 variants of research interest across BRAF, EGFR, ERBB2, KRAS, and PIK3CA
- Detection of variants at allele frequencies as low as 1%
- Use of as little as 10 ng of DNA
- Approximately 1 hour of hands-on time and 8 hours total time

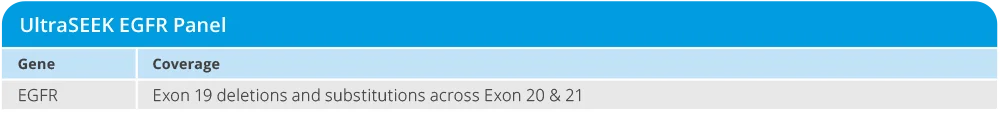
The UltraSEEK EGFR panel enables variant detection of EGFR markers in non-small cell lung cancer research samples, featuring:
- Identification of six EGFR variants of research interest, including T790M, C797S, E746_A750del, and L858R
- Detection of variants at allele frequencies as low as 0.1%
- Use of as little as 10 ng of cfDNA
- Approximately 1 hour of hands-on time and 8 hours total time
The UltraSEEK Colon panel enables multiplexed variant detection in colorectal cancer research, featuring:
- Identification of more than 80 variants of research interest across BRAF, EGFR, KRAS, NRAS, and PIK3CA
- Detection of variants at allele frequencies as low as 0.1%
- Use of as little as 10 ng of cfDNA
- Approximately 1 hour of hands-on time and 8 hours total time

The UltraSEEK Melanoma panel enables multiplexed variant detection in melanoma research samples, featuring:
- Identification of more than 60 variants of research interest across 13 genes, including BRAF, KIT, and NRAS
- Detection of variants at allele frequencies as low as 0.1%
- Use of as little as 10 ng of cfDNA
- Approximately 1 hour of hands-on time and 8 hours total time
Custom Panels
In addition to the panels mentioned above, Agena Bioscience also offers custom research panels unique to individual customer needs through Assays by Agena Laboratory Services. The iPLEX Pro, iPLEX HS, UltraSEEK, and Agena ClearSEEK chemistries are available for use in these custom assays, which are developed in coordination with each laboratory’s research needs.
References:
1. Gabriel S, Ziaugra L, Tabbaa D. (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet 60:2.12.1-2.12.18. 2. AH Box et al. Evaluation of a Mass Spectrometry-Based PIK3CA Mutation Assay for Predictive Breast Cancer Therapeutic Decision Making. Poster session presented at: Association of Molecular Pathology Annual Meeting; 2020. 3. National Comprehensive Cancer Network (NCCN) Guidelines – Breast Cancer. Version 2.2022.