Introduction
 Molecular genetic research continues to advance our understanding of complex biological systems. One of the key enabling technologies is mass spectrometry, which allows scientists to precisely identify and measure the presence of certain molecules in research samples. This approach plays a central role in pharmacogenomics (PGx) research, mutation profiling, genotyping SNPs and indels, copy number variation (CNV) analysis, and biomarker detection.
Molecular genetic research continues to advance our understanding of complex biological systems. One of the key enabling technologies is mass spectrometry, which allows scientists to precisely identify and measure the presence of certain molecules in research samples. This approach plays a central role in pharmacogenomics (PGx) research, mutation profiling, genotyping SNPs and indels, copy number variation (CNV) analysis, and biomarker detection.
The MassARRAY® System by Agena Bioscience® (RUO) utilizes mass spectrometry across multiple applications in genomics research. Its unique chemistries enable high-throughput workflows and provide benefits over alternatives such as real-time PCR and next-generation sequencing (NGS) for PGx research, variant identification, and custom assay development.
Topics
- Understanding MALDI-TOF technology
- Applications for MALDI-TOF technology in genomics research
- How the chemistry of the MassARRAY System sets the stage for MALDI-TOF
- How the MassARRAY System utilizes MALDI-TOF and unique chemistry to identify genetic variation
- Key benefits of the MassARRAY for molecular research
- Different applications for the MassARRAY System
Understanding MALDI-TOF Technology
The basic stages of mass spectrometry (MS) are ionization, acceleration, deflection, and detection. During a classic MS process, the ionization stage happens when the sample is vaporized and then passes into an ionization chamber, resulting in positively charged ions. During the acceleration stage, the positively charged ionization chamber repels the positively charged ions, accelerating them towards three negatively charged slits with progressively decreasing voltage. The speed at which the ions accelerate depends on their mass, where lighter ions move faster than heavier ones.
In the deflection stage, the stream of positively charged ions is deflected by a magnetic field. The deflection depends on the ion’s mass and charge, with stronger deflection occurring with ions of less mass or with a greater charge. In the final detection stage, the ions are detected by a sensor based on m/z. When an ion hits the detector, it generates an electrical current proportional to its abundance. The signal data are captured as a mass spectrum, and the m/z values are analyzed.
With “time of flight” (TOF) MS, the process accelerates ionized samples of different masses and charges to the same energy using an electric field before separating and detecting the ions based on their travel time through a vacuum flight tube of defined length. Because ions with different m/z have different initial velocities, and those with less mass and/or with more charge “fly” faster, each hits the detector at a slightly different time. Each impact with the detector generates an electrical signal, and those data are calculated and displayed as a mass spectrum of the different ion species in the sample.
The ionization method was improved with the invention of matrix-assisted laser desorption/ionization (MALDI). This improved ionization method does not lead to a significant loss of sample integrity, which is important for fragile compounds and large molecules. The combination of MALDI ionization and TOF detection gives us the MALDI-TOF Mass Spectrometry widely used in genomics research today.
Applications for MALDI-TOF technology in genomics research
MALDI-TOF MS has been applied to detect molecules ranging from nucleic acids and proteins to amino acids and lipids. In recent years, its research applications have expanded to:
- PGx research: Detecting SNPs, indels, and CNVs in pharmacogenomic panels, including CYP2D6 hybrid alleles and other CYP450 family genes.
- Mutational profiling: Somatic variant identification in research samples and studies of circulating nucleic acids for liquid biopsy research
- Molecular biology applications: Protein biomarker analysis, small molecule identification, and epidemiological studies.
From 2018–2022, “nucleic acids analysis” and “research into disease mechanisms” were among the most frequently co-occurring terms with MALDI-TOF in published reviews, underscoring its importance in genomics, molecular research, and biomarker identification.
How the chemistry of the MassARRAY System sets the stage for MALDI-TOF
MALDI-TOF is the technology used for the detection of nucleic acid samples on the MassARRAY System (RUO), an automated platform by Agena Bioscience. Before analysis, the samples are prepared using specific panel chemistries to detect genetic variations such as single nucleotide polymorphisms (SNPs), insertions/deletions (indels), and copy number variation (CNV). The base chemistry uses multiplexed PCR (Polymerase Chain Reaction) in combination with primer extension.
The most common sample preparation method used for SNP genotyping with MALDI-TOF MS includes the following steps:
Extraction
MassARRAY panel chemistries require DNA or RNA extracted as the first input sample. Suitable DNA sample sources include FFPE tissue, fine needle aspirates, buccal cells, or blood plasma, depending on which genetic research panel is used. The DNA/RNA extraction process can be completed using one of many off-the-shelf extraction kits and systems that are widely available. Extraction isolates the DNA or RNA in the sample and removes other potentially interfering substances. The extracted DNA or RNA is then transferred to a 96- or 384-well microtiter plate.
Conventional PCR
After extraction, target DNA or RNA sequences are amplified by conventional PCR using multiplexed panel-specific primers. The resulting amplicons are typically about 60–800 bp in length and include sites of interest for genetic variations. Up to 100 primers (for 50 segments) can be amplified in a single reaction using only 5 ng of input sample DNA.
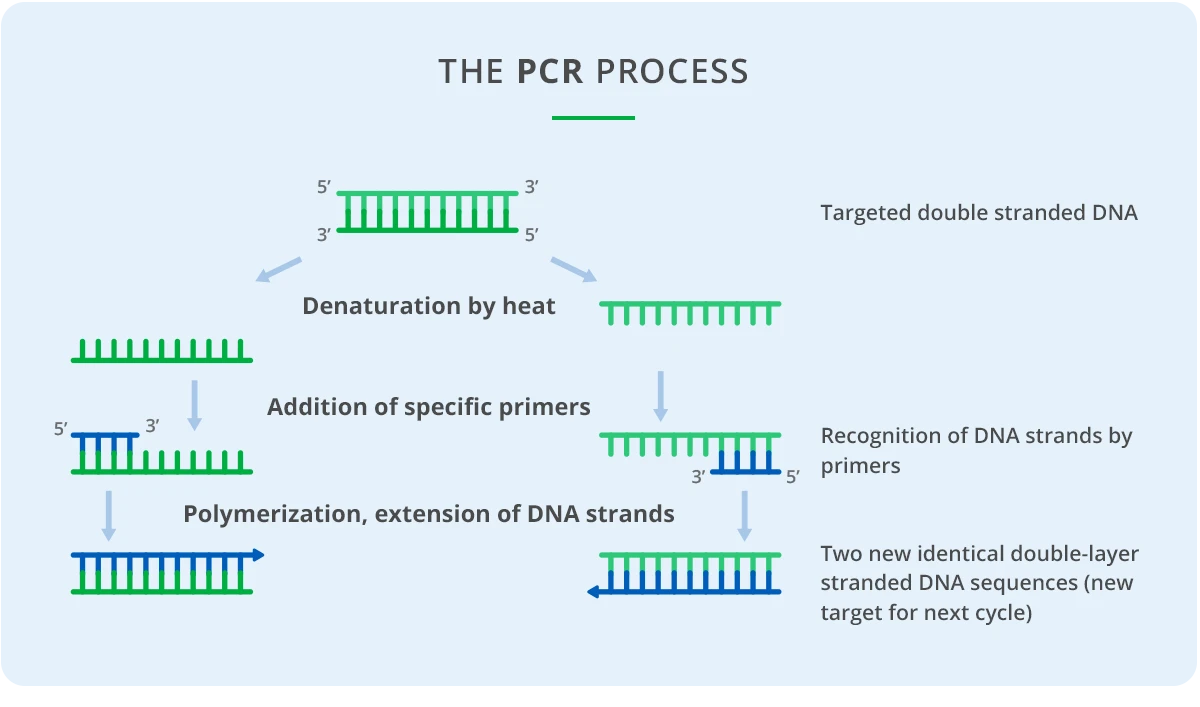
Purification
After PCR amplification of the target genes is complete, dNTPs remaining in the amplification reaction mixture are dephosphorylated by an enzyme called shrimp alkaline phosphatase (SAP). This is critical because the nucleotides need to be inactivated so that they cannot be incorporated in the next step, single-base extension.
Single Base Extension
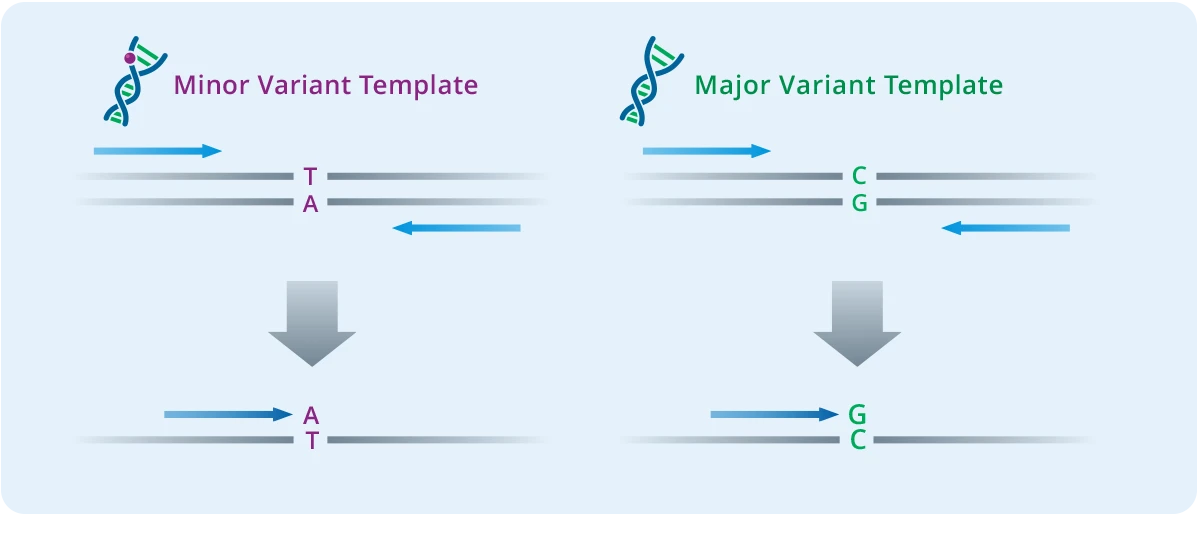
The single-base extension step creates the analyte for the MALDI-TOF analysis. This step uses panel-specific extension primers, and termination mixes to generate DNA products with different masses depending on which genetic variants are present. The extension primers anneal next to each variant site being assayed, and the extension reaction incorporates one base that is a dideoxy A, T, C, or G terminator nucleotide. Because each nucleotide has a different mass, extension products with even just one base difference can be distinguished by MS.
Multiple primers can be multiplexed in a single reaction so long as their masses are different from the other primer extension product combinations in the same well. This results in an extremely high level of multiplexing of up to 50 primers per well. The final product is then diluted with water to ensure there is enough sample volume for analysis.
How the MassARRAY System utilizes MALDI-TOF and unique chemistry to identify genetic variation
The reaction products move onto the MassARRAY System for final sample preparation and analysis. The Chip Prep Module (CPM) finalizes sample preparation for mass spectrometry and transfers the analytes to the MassARRAY Analyzer 4 (MA4). The reaction products are desalted and transferred to a SpectroCHIP®, a silicon chip pre-spotted with MALDI matrix.
 The analyte and matrix co-crystallize upon drying, and the chip is automatically moved into the MA4 for analysis. Once in the MS, each pad on the chip array is irradiated with laser pulses to desorb and ionize the analyte-matrix crystals. The resulting positively charged DNA molecules accelerate up the mass spectrometer flight tube toward the detector. Separation occurs by time-of-flight, with the lightest ions reaching the detector first.
The analyte and matrix co-crystallize upon drying, and the chip is automatically moved into the MA4 for analysis. Once in the MS, each pad on the chip array is irradiated with laser pulses to desorb and ionize the analyte-matrix crystals. The resulting positively charged DNA molecules accelerate up the mass spectrometer flight tube toward the detector. Separation occurs by time-of-flight, with the lightest ions reaching the detector first.
The time-of-flight data are processed by MassARRAY Typer software, which differentiates the extension products by mass and generates variant summary reports for research applications, including DNA variant calls for PGx panels, oncology research, and CNV detection.
Key benefits of the MassARRAY System for molecular research
The MassARRAY System (RUO) is a powerful tool for molecular genetic research, offering several benefits over more conventional methods such as real-time PCR and next-generation sequencing (NGS):
- High Multiplexing: Analyze up to 50 targets per well, enabling efficient PGx research and variant profiling
- Low Sample Input: Requires minimal DNA, making it suitable for liquid biopsy research and degraded FFPE samples.
- Cost-Effective: Provides cost savings compared to NGS by avoiding expensive fluorescent dyes and complex bioinformatics.
- Rapid Turnaround: From DNA extraction to data, under a single research day.
Different applications for the MassARRAY System
The MassARRAY System is a powerful tool for genetic research, offering high levels of multiplexing and cost-effective, high-throughput research. It has several key applications, including pharmacogenetics (PGx) research, mutation profiling, and sample identification.
- Pharmacogenomics (PGx research): Identify SNPs, indels, CNVs, and hybrid alleles across genes such as CYP2D6 to study drug metabolism and variability.
- Mutation Profiling: Identify sequence variants in research samples, including circulating tumor DNA (ctDNA) through liquid biopsy research.
- Sample identification: Ensure sample integrity through DNA fingerprinting to reduce research errors.
In addition to these applications, Assays by Agena Laboratory Services offer custom multiplex assays using markers of interest across many application areas. These custom panels also use the MassARRAY System workflow. Overall, the MassARRAY System is a versatile tool with a wide range of applications in genetic research.
References:
1. Everley, R. A., Mott, T. M., Wyatt, S. A., Toney, D. M., and Croley, T. R. (2008). Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J. Am. Soc. Mass Spectrom. 19, 1621–1628. doi: 10.1016/j.jasms.2008.07.003 2. Ekström, S., Onnerfjord, P., Nilsson, J., Bengtsson, M., Laurell, T., and Marko-Varga, G. (2000). Integrated microanalytical technology enabling rapid and automated protein identification. Anal. Chem. 72, 286–293. doi: 10.1021/ac990731l 3. De Ranieri, E. When a velocitron meets a reflectron. Nature Methods 4, 8 (2015). doi:10.1038/nmeth.3526 4. Cornish, Timothy & Bryden, Wayne. (1999). Miniature Time-of-Flight Mass Spectrometer for a Field-Portable Biodetection System. Johns Hopkins APL Tech Digest. 20. 5. Dandan Li, Jia Yi, Guobin Han, and Liang Qiao, MALDI-TOF Mass Spectrometry in Clinical Analysis and Research, ACS Measurement Science Au 2022 2 (5), 385-404, DOI: 10.1021/acsmeasuresciau.2c00019 6. Amanda Rae Buchberger, Kellen DeLaney, Jillian Johnson, and Lingjun Li, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights, Analytical Chemistry 2018 90 (1), 240-265, DOI: 10.1021/acs.analchem.7b04733 8. Vogel N, Schiebel K, Humeny A. Technologies in the Whole-Genome Age: MALDI-TOF-Based Genotyping. Transfus Med Hemother. 2009;36(4):253-262. doi: 10.1159/000225089. Epub 2009 Jul 10. PMID: 21049076; PMCID: PMC2941830. 9. Li, D., Yi, J., Han, G., & Qiao, L. (2022). MALDI-TOF Mass Spectrometry in Clinical Analysis and Research. ACS Measurement Science Au, 2(5), 385-404. 10. Newborn Screening Ontario, Ottawa, ON, NSO Symposium 2015 Poster.